Publications
Please note that my publications are under my legal name ‘Shachi Deshpande’.
2025
-
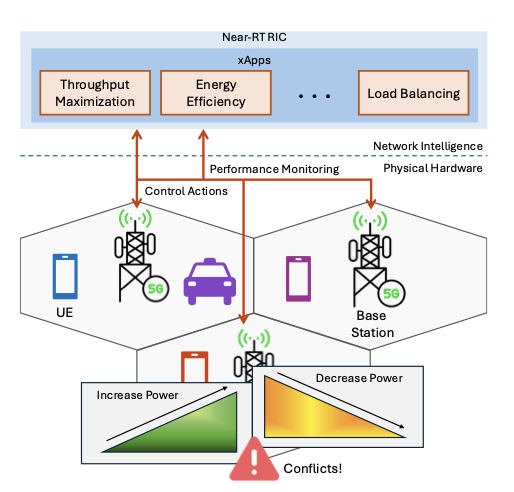 Towards xApp Conflict Evaluation with Explainable Machine Learning and Causal Inference in O-RANPragya Sharma, Shihua Sun, Shachi Deshpande, and 2 more authorsIn Proceedings of the IEEE Conference on Network Function Virtualization and Software Defined Networks (NFV-SDN) , 2025Best Paper Award
Towards xApp Conflict Evaluation with Explainable Machine Learning and Causal Inference in O-RANPragya Sharma, Shihua Sun, Shachi Deshpande, and 2 more authorsIn Proceedings of the IEEE Conference on Network Function Virtualization and Software Defined Networks (NFV-SDN) , 2025Best Paper AwardThe Open Radio Access Network (O-RAN) architecture enables a flexible, vendor-neutral deployment of 5G networks by disaggregating base station components and supporting third-party xApps for near real-time RAN control. However, the concurrent operation of multiple xApps can lead to conflicting control actions, which may cause network performance degradation. In this work, we propose a framework for xApp conflict management that combines explainable machine learning and causal inference to evaluate the causal relationships between RAN Control Parameters (RCPs) and Key Performance Indicators (KPIs). We use model explainability tools such as SHAP to identify RCPs that jointly affect the same KPI, signaling potential conflicts, and represent these interactions as a causal Directed Acyclic Graph (DAG). We then estimate the causal impact of each of these RCPs on their associated KPIs using metrics such as Average Treatment Effect (ATE) and Conditional Average Treatment Effect (CATE). This approach offers network operators guided insights into identifying conflicts and quantifying their impacts, enabling more informed and effective conflict resolution strategies across diverse xApp deployments.
-
 Calibrated Regression Against An Adversary Without RegretShachi Deshpande, Charles Marx, and Volodymyr KuleshovIn Proceedings of the Forty-first Conference on Uncertainty in Artificial Intelligence , 21–25 jul 2025
Calibrated Regression Against An Adversary Without RegretShachi Deshpande, Charles Marx, and Volodymyr KuleshovIn Proceedings of the Forty-first Conference on Uncertainty in Artificial Intelligence , 21–25 jul 2025We are interested in probabilistic prediction in online settings in which data does not follow a probability distribution. Our work seeks to achieve two goals: (1) producing valid probabilities that accurately reflect model confidence; (2) ensuring that traditional notions of performance (e.g., high accuracy) still hold. We introduce online algorithms guaranteed to achieve these goals on arbitrary streams of data-points, including data chosen by an adversary. Specifically, our algorithms produce forecasts that are (1) calibrated i.e., an 80% confidence interval contains the true outcome 80% of the time-and (2) have low regret relative to a user-specified baseline model. We implement a post-hoc recalibration strategy that provably achieves these goals in regression; previous algorithms applied to classification or achieved (1) but not (2). In the context of Bayesian optimization, an online model-based decision-making task in which the data distribution shifts over time, our method yields accelerated convergence to improved optima.
2024
-
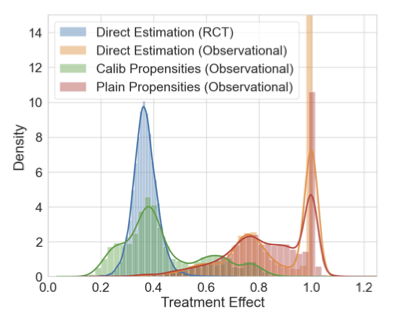 Calibrated Propensity Scores for Causal Effect EstimationShachi Deshpande, and Volodymyr KuleshovIn Uncertainty in Artificial Intelligence (UAI) , 21–25 jul 2024
Calibrated Propensity Scores for Causal Effect EstimationShachi Deshpande, and Volodymyr KuleshovIn Uncertainty in Artificial Intelligence (UAI) , 21–25 jul 2024Propensity scores are commonly used to estimate treatment effects from observational data. We argue that the probabilistic output of a learned propensity score model should be calibrated – i.e., a predictive treatment probability of 90% should correspond to 90% of individuals being assigned the treatment group – and we propose simple recalibration techniques to ensure this property. We prove that calibration is a necessary condition for unbiased treatment effect estimation when using popular inverse propensity weighted and doubly robust estimators. We derive error bounds on causal effect estimates that directly relate to the quality of uncertainties provided by the probabilistic propensity score model and show that calibration strictly improves this error bound while also avoiding extreme propensity weights. We demonstrate improved causal effect estimation with calibrated propensity scores in several tasks including high-dimensional image covariates and genome-wide association studies (GWASs). Calibrated propensity scores improve the speed of GWAS analysis by more than two-fold by enabling the use of simpler models that are faster to train.
-
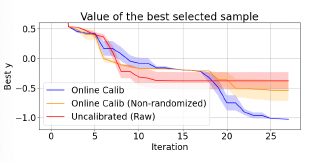
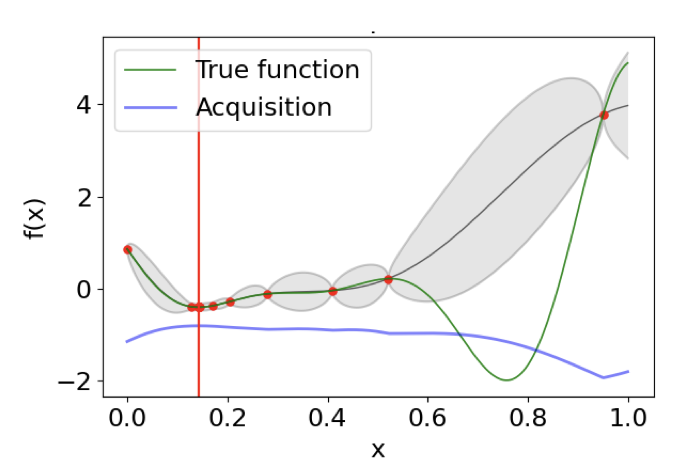 Online Calibrated and Conformal Prediction Improves Bayesian OptimizationShachi Deshpande, Charles Marx, and Volodymyr KuleshovIn Artificial Intelligence and Statistics (AISTATS) , 21–25 jul 2024
Online Calibrated and Conformal Prediction Improves Bayesian OptimizationShachi Deshpande, Charles Marx, and Volodymyr KuleshovIn Artificial Intelligence and Statistics (AISTATS) , 21–25 jul 2024Accurate uncertainty estimates are important in sequential model-based decision-making tasks such as Bayesian optimization. However, these estimates can be imperfect if the data violates assumptions made by the model (e.g., Gaussianity). This paper studies which uncertainties are needed in model-based decision-making and in Bayesian optimization, and argues that uncertainties can benefit from calibration – i.e., an 80% predictive interval should contain the true outcome 80% of the time. Maintaining calibration, however, can be challenging when the data is non-stationary and depends on our actions. We propose using simple algorithms based on online learning to provably maintain calibration on non-i.i.d. data, and we show how to integrate these algorithms in Bayesian optimization with minimal overhead. Empirically, we find that calibrated Bayesian optimization converges to better optima in fewer steps, and we demonstrate improved performance on standard benchmark functions and hyperparameter optimization tasks.
2022
-
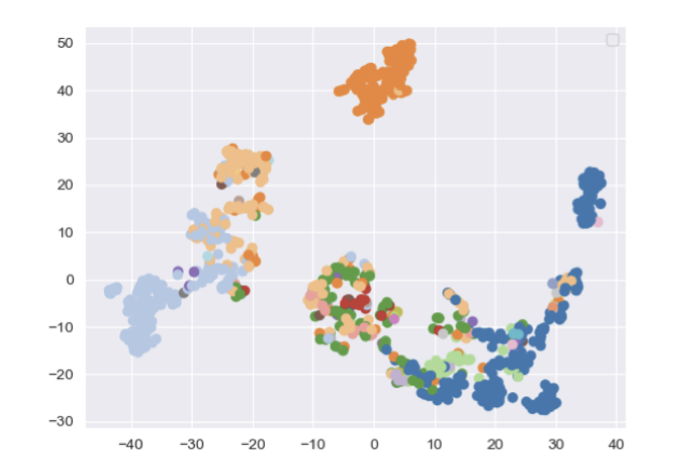 Deep Multi-Modal Structural Equations For Causal Effect Estimation With Unstructured ProxiesShachi Deshpande, Kaiwen Wang, Dhruv Sreenivas, and 2 more authorsIn Advances in Neural Information Processing Systems , 21–25 jul 2022
Deep Multi-Modal Structural Equations For Causal Effect Estimation With Unstructured ProxiesShachi Deshpande, Kaiwen Wang, Dhruv Sreenivas, and 2 more authorsIn Advances in Neural Information Processing Systems , 21–25 jul 2022Estimating the effect of intervention from observational data while accounting for confounding variables is a key task in causal inference. Oftentimes, the confounders are unobserved, but we have access to large amounts of additional unstructured data (images, text) that contain valuable proxy signal about the missing confounders. This paper argues that leveraging this unstructured data can greatly improve the accuracy of causal effect estimation. Specifically, we introduce deep multi-modal structural equations, a generative model for causal effect estimation in which confounders are latent variables and unstructured data are proxy variables. This model supports multiple multi-modal proxies (images, text) as well as missing data. We empirically demonstrate that our approach outperforms existing methods based on propensity scores and corrects for confounding using unstructured inputs on tasks in genomics and healthcare. Our methods can potentially support the use of large amounts of data that were previously not used in causal inference
-
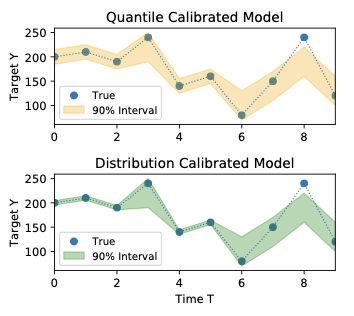 Calibrated and Sharp Uncertainties in Deep Learning via Density EstimationVolodymyr Kuleshov, and Shachi DeshpandeIn International Conference on Machine Learning, ICML 2022, 17-23 July 2022, Baltimore, Maryland, USA , 21–25 jul 2022
Calibrated and Sharp Uncertainties in Deep Learning via Density EstimationVolodymyr Kuleshov, and Shachi DeshpandeIn International Conference on Machine Learning, ICML 2022, 17-23 July 2022, Baltimore, Maryland, USA , 21–25 jul 2022Accurate probabilistic predictions can be characterized by two properties – calibration and sharpness. However, standard maximum likelihood training yields models that are poorly calibrated and thus inaccurate – a 90% confidence interval typically does not contain the true outcome 90% of the time. This paper argues that calibration is important in practice and is easy to maintain by performing low-dimensional density estimation. We introduce a simple training procedure based on recalibration that yields calibrated models without sacrificing overall performance; unlike previous approaches, ours ensures the most general property of distribution calibration and applies to any model, including neural networks. We formally prove the correctness of our procedure assuming that we can estimate densities in low dimensions and we establish uniform convergence bounds. Our results yield empirical performance improvements on linear and deep Bayesian models and suggest that calibration should be increasingly leveraged across machine learning.
2017
-
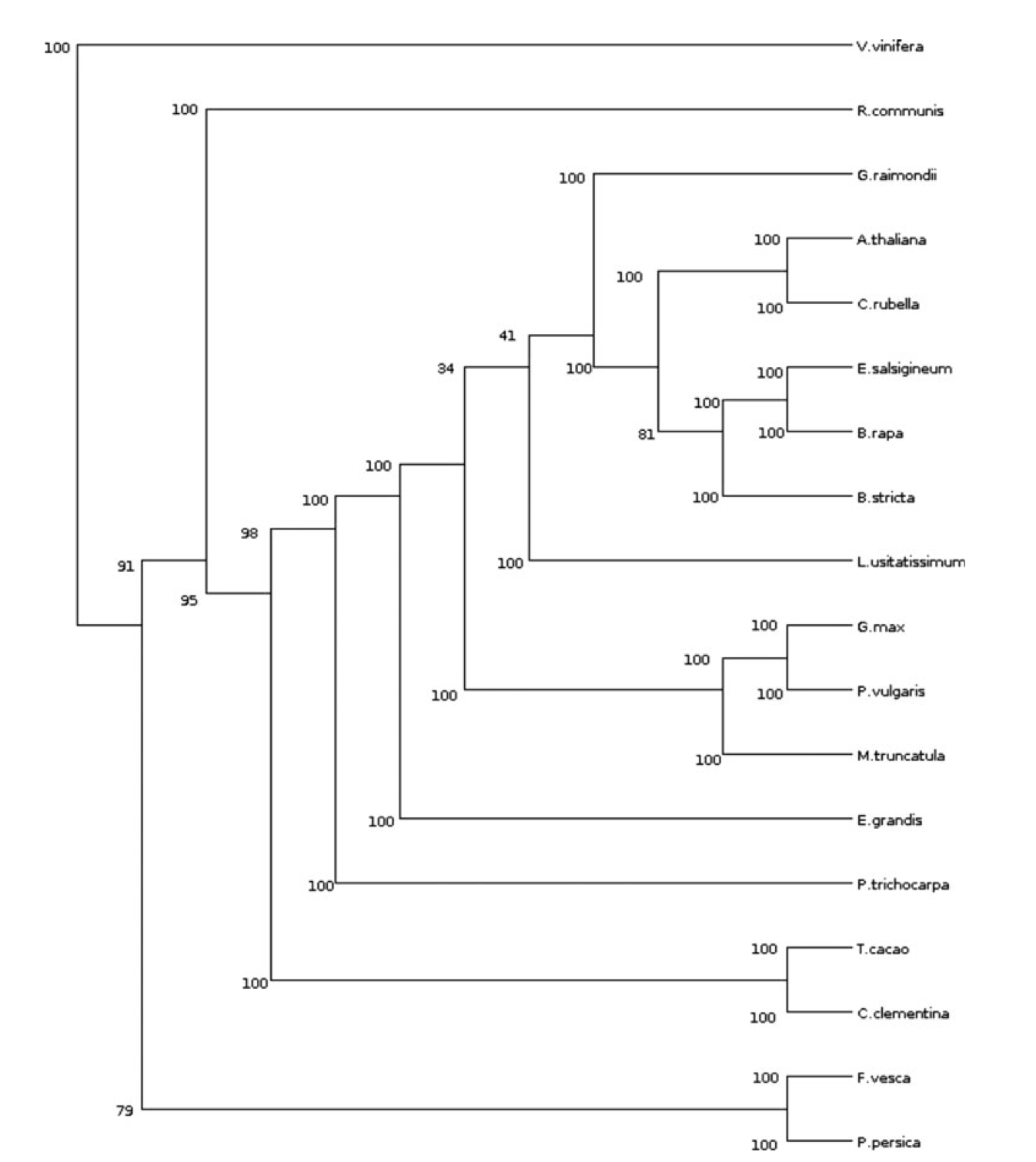 New Genome Similarity Measures based on Conserved Gene AdjacenciesDaniel Doerr, Luis Antonio Brasil Kowada, Eloi Araujo, and 4 more authorsJ. Comput. Biol., 21–25 jul 2017
New Genome Similarity Measures based on Conserved Gene AdjacenciesDaniel Doerr, Luis Antonio Brasil Kowada, Eloi Araujo, and 4 more authorsJ. Comput. Biol., 21–25 jul 2017Many important questions in molecular biology, evolution, and biomedicine can be addressed by comparative genomic approaches. One of the basic tasks when comparing genomes is the definition of measures of similarity (or dissimilarity) between two genomes, for example, to elucidate the phylogenetic relationships between species. The power of different genome comparison methods varies with the underlying formal model of a genome. The simplest models impose the strong restriction that each genome under study must contain the same genes, each in exactly one copy. More realistic models allow several copies of a gene in a genome. One speaks of gene families, and comparative genomic methods that allow this kind of input are called gene family-based. The most powerful-but also most complex-models avoid this preprocessing of the input data and instead integrate the family assignment within the comparative analysis. Such methods are called gene family-free. In this article, we study an intermediate approach between family-based and family-free genomic similarity measures. Introducing this simpler model, called gene connections, we focus on the combinatorial aspects of gene family-free genome comparison. While in most cases, the computational costs to the general family-free case are the same, we also find an instance where the gene connections model has lower complexity. Within the gene connections model, we define three variants of genomic similarity measures that have different expression powers. We give polynomial-time algorithms for two of them, while we show NP-hardness for the third, most powerful one. We also generalize the measures and algorithms to make them more robust against recent local disruptions in gene order. Our theoretical findings are supported by experimental results, proving the applicability and performance of our newly defined similarity measures.